|
You probably haven’t heard of the
virus, known as
human respiratory syncytial virus,
but you probably have had experience with it. Respiratory syncytial virus, RSV for short, is so common that almost every child in the United States under two years of age has been infected once, and that half of children under three have been infected at least
twice.
Symptoms are similar to a common cold—runny nose, coughing, wheezing, perhaps a fever—and usually clear up within two weeks.
In young children, though, RSV infection can be more severe, causing infections of the lower
respiratory tract
that require hospitalization. In fact, RSV is the leading cause of lower respiratory infections in young children in the United States, resulting in an estimated 75,000 to 125,000 hospitalizations
anually.
Furthermore, young children who contract a serious RSV infection are much more likely to develop
asthma
and other respiratory problems in the future. To date, available treatments can only address the symptoms of infection while the virus runs its course.
Could we know more?
Given the number of people, particularly young children, affected by the virus, it is surprising how little we know about it. One possibility of how RSV infection may lead to asthma is that the virus hangs around in the lungs even after symptoms have disappeared. As a result of this “viral persistence,” years later the same virus may trigger the symptoms we know as asthma. Another possibility is that RSV infection may trigger asthma in children who were already genetically predisposed to it.
Dr. Richard Hegele at the University of Toronto in Canada has spent his career studying the respiratory syncytial virus and his team has contributed to our knowledge of the links between RSV and asthma. His latest research provides insight into the workings of this elusive virus that may soon lead to the development of new ways to prevent RSV infection in the first place.
Key question: How does the virus get in?
The most basic question for Dr. Hegele was how RSV gains entry into the cells in order to begin an infection. This is important because viruses need to enter the cell to reproduce. Dr. Hegele and his team hypothesized that the virus would bind to a component on the cell surface, known as a
receptor,
in order to gain access to the cell. The cell surface has three major parts – protein, fat, or carbohydrate. Dr. Hegele and his team hypothesized that RSV bound to one or more of these components.
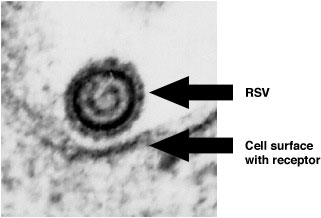
High Magnification Electron Micrograph of RSV Near the Cell Surface
To figure out which one, Dr. Hegele and his team prepared the cell surface in such a way that it lacked one of these three components. This is done using special proteins called
enzymes
that will break down one component. For example, the cells treated with an enzyme to break down proteins would lack proteins on the cell surface. This process was repeated with carbohydrates and fats. Dr. Hegele and his team then infected the different cell preparations with RSV. They found that RSV was able to infect the cells that lacked fats and sugars on the cell surface, but not those that lacked proteins.
From this result, Dr. Hegele concluded that RSV interacted with some protein on the cell surface to gain entry into the cell.
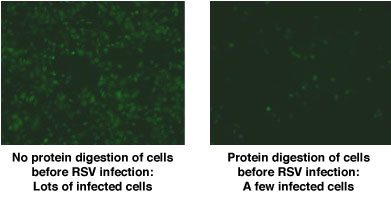
Low Magnification Fluorescence Microscopy: RSV-Infected Cells are Green
Conclusion: RSV receptor contains protein
One of thousands
The next step was to figure out which particular protein allowed RSV into the cell. There are thousands of proteins on the cell surface, so isolating a few candidates was a challenge. Dr. Hegele began by using a method called Viral Overlay Protein Binding Assay that exposed different cell-surface proteins to RSV. In order to see where the virus stuck, the researchers used proteins called
antibodies
that bind only with RSV. From these experiments, Dr. Hegele and his team narrowed the field of thousands of proteins down to just a few dozen.
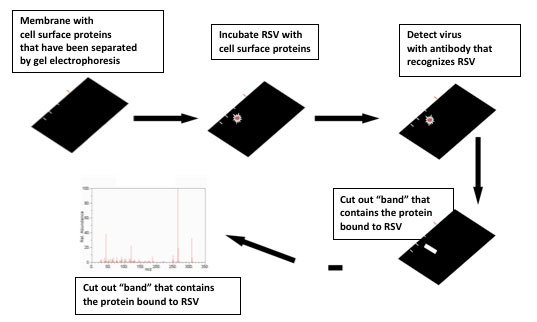
Viral Overlay Protein Binding Assay
Next, the researchers used
mass spectrometry
to determine the identity of each of the proteins that bound RSV. Mass spectrometry works by separating an unknown compound into charged particles that accelerate at a certain rate based on their mass and charge. Every compound has a distinct pattern when visualized using mass spectrometry. The unknown proteins were identified by matching their mass spectrometry images with those of a library of known compounds.
Dr. Hegele and his team did this entire process not only in human cells but in hamster and dog cells as well. They found that a protein called
nucleolin
bound to RSV in all three types of cells. As a result, the researchers hypothesized that nucleolin was the candidate RSV receptor on the cell surface they had been looking for. Next, these researchers performed four kinds of experiments to verify that nucleolin was the RSV receptor.

RSV Viral Overlay Protein Binding Assay
Verification
It is not uncommon in scientific research to use many approaches when trying to validate a result. Though it slows things down, this produces far more reliable and reproducible results.
First, the researchers used a method called competition. Here’s the logic: if nucleolin is the RSV receptor, then if you introduce another molecule that will bind to nucleolin in competition with RSV, fewer cells should get infected. The researchers took a
petri dish
full of cells and treated them not only with RSV but also with antibodies that would bind to nucleolin – the competition. When they compared this petri dish to a control dish treated only with RSV, they found that many fewer cells were infected. This is exactly what you would expect if nucleolin was the RSV receptor, and these results supported the team’s hypothesis.
In a second set of experiments, researchers bought nucleolin (from a special chemical supplier) and put it in a liquid solution with RSV. They then treated cells with the virus alone, with nucleolin alone and with nucleolin and bound RSV. They found that in the nucleolin and bound RSV petri dishes, fewer cells became infected. This was expected since some of the virus had already bound to nucleolin in the solution. These experiments provided more evidence that nucleolin was the RSV receptor.
Next, Dr. Hegele and his team used small molecules called
small interfering RNA
(siRNA) that prevent the production of nucleolin in cells. When cells that lacked nucleolin as a result of treatment with siRNAs were then exposed to the virus, they weren’t infected. This was even further proof that nucleolin was indeed the RSV receptor the researchers were looking for.
Lastly, the researchers did the opposite experiment using a type of cell that cannot be infected by RSV because it lacks nucleolin on the cell surface. Finding such a type of cell was difficult, since RSV can infect many types of animal cells. In a literature search on the subject, though, the research team discovered a type of moth that was resistant to RSV infection. One of the team members had worked with the moth cells before, and was able to set up an experiment that introduced the human gene for nucleolin into the moth DNA. When the moth cells that had the human nucleolin gene were exposed to RSV, they became infected. Taken together, these results allowed the researchers to conclude with great certainty that nucleolin was indeed the RSV receptor on the cell surface.
Going live
Up until now, all the experiments had been done with cells in petri dishes. The next step was to determine whether similar results could be obtained in animal models. To do this, the researchers used siRNAs to prevent the production of nucleolin on the cell surface of lung cells in mice. When the treated mice were then exposed to the virus, they did not become infected as the controls did. “This sealed the deal,” remarked Dr. Hegele. “We now know that in the body, in the lungs, nucleolin is functioning as an RSV receptor.”
These results are very exciting because they provide a new target for preventing RSV infection. Dr. Hegele’s ongoing research focuses on understanding which part of the protein the virus binds to in order to make even more effective therapies. He also wants to know whether nucleolin appears on the cell surface in the same way in all diseases. In particular, he is curious whether patients with asthma or other lung diseases have more or less nucleolin on the surfaces of their lung cells than healthy patients. Dr. Hegele and his team continue to collaborate with people from all over the world to work towards answering these questions.
Dr. Richard Hegele is a Professor of Laboratory Medicine and
Pathobiology
at the University of Toronto. His research focuses on the human respiratory syncytial virus, how it causes infection, and potential therapeutic options. When not in the lab, Dr. Hegele enjoys reading, listening to music, and spending time outdoors.
For More Information:
- Tayyari, F. et al. 2011. “Identification of nucleolin as a cellular receptor for human respiratory syncytial virus.” Nature Medicine, 17(9): 1132-1136.
To Learn More:
Rebecca Kranz with Andrea Gwosdow, PhD Gwosdow Associates
HOME | ABOUT | ARCHIVES | TEACHERS | LINKS | CONTACT
All content on this site is © Massachusetts
Society for Medical Research or others. Please read our copyright
statement — it is important. |

