| |
On June 7, 2012, the Centers for Disease Control and Prevention (CDC) reported that 123 people in 25 states had been infected with a strain of Salmonella bacteria traced to chicks and ducklings from a hatchery in Ohio. That same month, 390 people in 27 states were infected with Salmonella traced to frozen tuna, and an additional 22 people in 13 states were infected from dry dog food. In fact, approximately 1 million people are infected with Salmonella annually in the United Sates alone, and nearly 400 people died in 2006 from Salmonella-related illness . In 2010, Salmonella was the most common foodborne infection reported.
On an international level, Salmonella food poisoning is even more devastating; at least 80 million people are infected annually, resulting in approximately 155,000 deaths.
There are over 2,000 different strains of Salmonella bacteria that cause infections ranging from mild food poisoning to life-threatening Typhoid fever. The bacteria can be associated with raw eggs, meat, and dairy products, as well as vegetables such as tomatoes, lettuce or spinach.
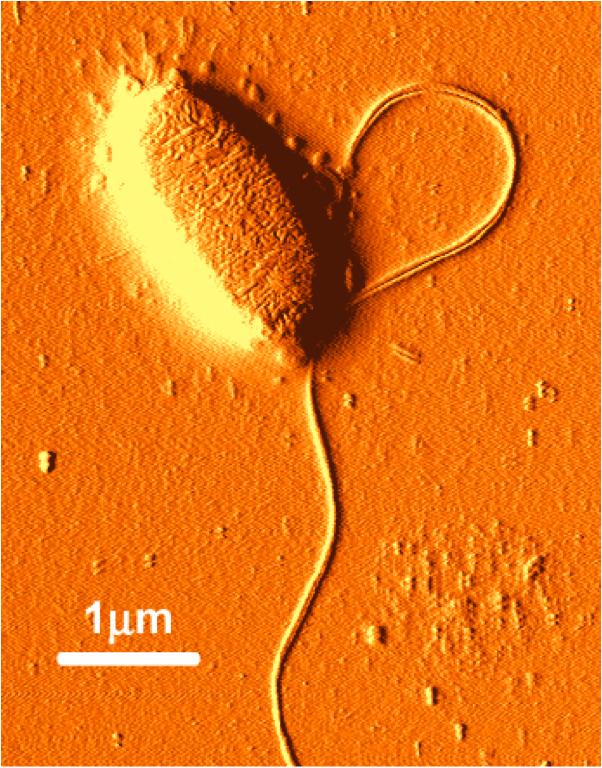
Salmonella typhimurium – photo courtesy of Institute of Food Research, UK
Patients infected with Salmonella bacteria will usually experience diarrhea, fever, and abdominal cramps within 12 to 72 hours. Symptoms usually last 4 to 7 days and most people recover without treatment.
In some cases, though, the diarrhea can become so severe to warrant hospitalization, and many deaths occur in parts of the world where such treatments are not available. A particularly nasty strain of Salmonella, called Salmonella typhi causes the life-threatening typhoid fever. Most cases of typhoid in the United States are contracted internationally, usually because of poor hygiene. Worldwide, typhoid affects over 21 million people annually.
Salmonella infection remains a huge healthcare issue in the United States and internationally because the bacteria are able to adapt so quickly to many environmental extremes. From the skin of a tomato or a raw piece of meat, an ingested bacterium is first exposed to the acidic stomach juices that kill most other bacteria, but not Salmonella.
The Salmonella next enters the intestines where it survives the salty, oxygen-poor environment to settle into the intestinal lining and cause infection. “We desperately need a vaccine and better antibiotics to combat Salmonella infection and save lives,” comments Professor Jay Hinton. “But to do that, we first need to understand what makes Salmonella so good at infecting people.”
Jay Hinton is a Professor of Microbial Pathogenesis at the University of Liverpool, UK, and has spent most of his career attempting to do just that. Recently he and his team have discovered how Salmonella bacteria regulate their genes so quickly to adapt to new environments. The researchers hope that their findings will speed the development of new treatments and preventative measures for Salmonella infection.
In the 1990’s, researchers discovered that the Salmonella typhimurium genome contained almost 5,000 genes. These genes provide the instructions for all the proteins the bacteria need to survive, thrive, and cause infection. In order to understand which genes were involved in infection, Professor Hinton’s lab used modern genetic techniques to analyze Salmonella throughout the disease process. They identified over 900 genes that are active during infection of mammalian cells.
“An impressive 1/5 of all Salmonella genes are involved in infection,” remarked Professor Hinton. “We wanted to know why.”
Subsequent research by Professor Hinton’s team and others around the world revealed how the bacteria are able to adapt so quickly to a variety of extreme environments—by modifying protein machines on the cell membrane to fit the environmental conditions. Salmonella are able to do this because they have many proteins and
small RNA molecules that choreograph the rapid switching of genes on and off, depending on their surroundings. In a particularly acidic environment, for example, the bacteria need different proteins to function on the cell surface than in a very salty environment. By turning on just the genes necessary for the particular conditions the bacteria are in, Salmonella can thrive in a huge variety of environments.
“The logical next step for our research was to identify how the genes that allow Salmonella to cause infection are controlled,” explained Professor Hinton. Using a variety of newly developed techniques, the research team identified over 1,800 genetic switches called promoters. “Thanks to my talented team, we have found the switches for genes that are needed for infection giving us an unprecedented view of the way that Salmonella cause human disease,” said Professor Hinton.
This knowledge is now being applied to a new type of Salmonella that is killing thousands of HIV-positive people in
Sub-Saharan Africa. This new strain of Salmonella typhimurium is the focus of Professor Hinton’s current work. “We are still years away from new treatments,” Professor Hinton acknowledged, “but these important findings will help us to ask why Salmonella has become so lethal in Africa.”
Jay Hinton is the Professor of Microbial Pathogenesis at the Institute of Integrative Biology in Liverpool, UK. He has spent his career studying Salmonella bacteria with the ultimate goal of developing preventative treatments for the disease. When not in the lab, Professor Hinton enjoys walking on beautiful beaches and sailing.
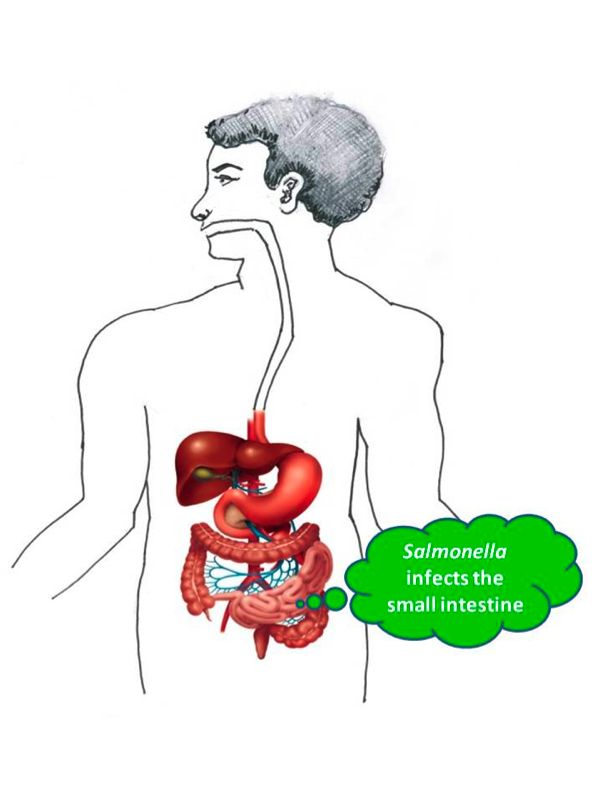
Salmonella at the small intestine
For More Information:
- Kroger, C. et al. 2012. “The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium.” Proceedings of the National Academy of Sciences. http://www.pnas.org/content/early/2012/04/24/1201061109.short and http://www.tcd.ie/Communications/news/pressreleases/pressRelease.php?headerID=2436&pressReleaseArchive=2012
- Eriksson, S. et al. 2003. “Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica.” Molecular Microbiology 47(1): 103-118. http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.2003.03313.x/full
To Learn More:
- US Centers for Disease Control and Prevention.
- World Health Organization.
- US Food and Drug Administration.
- US Department of Agriculture.
Rebecca Kranz with Andrea Gwosdow, PhD www.gwosdow.com
HOME | ABOUT | ARCHIVES | TEACHERS | LINKS | CONTACT
All content on this site is © Massachusetts
Society for Medical Research or others. Please read our copyright
statement — it is important. |
|
|
 Prof. Hinton and his lab team at rest
 Salmonella entering the intestinal tract
 Animation of Salmonella infection—narration by Prof. Brett Finlay
 “It’s amazing you’re not dead yet”—short lecture by Prof. Hinton
Sign Up for our Monthly Announcement!
...or  subscribe to all of our stories! subscribe to all of our stories!

What A Year! is a project of the Massachusetts
Society for Medical Research.
|
|

